My Grandmother mtDNA Haplogroup A2 Paternal Native American
My Grandmother mtDNA Haplogroup A2 Paternal Native American
mtDNA - Native American Maps
Welcome to
Clan Aiyana
Clan Aiyana
In human genetics, Haplogroup A is a human mitochondrial DNA (mtDNA) haplogroup.
A haplogroup is Believed to have Originated in Asia about 60.000 years before present. Its ancestral haplogroup was Haplogroup N.
A haplogroup is found throughout modern Asia. Its A1 subgroup is found in northern and central Asia, while its subgroup A2 is found in Siberia and is one of five haplogroups Also found nospovos indigenous of the Americas, the others being B, C, D and X.
The mummy "Juanita" from Peru, also called the "Ice Maiden", Has Been shown to belong to mitochondrial haplogroup A.
In his famous book The Seven Daughters of Eve, Bryan Sykes named the originator of this mitochondrial haplogroup Aiyana.
A haplogroup is Believed to have Originated in Asia about 60.000 years before present. Its ancestral haplogroup was Haplogroup N.
A haplogroup is found throughout modern Asia. Its A1 subgroup is found in northern and central Asia, while its subgroup A2 is found in Siberia and is one of five haplogroups Also found nospovos indigenous of the Americas, the others being B, C, D and X.
The mummy "Juanita" from Peru, also called the "Ice Maiden", Has Been shown to belong to mitochondrial haplogroup A.
In his famous book The Seven Daughters of Eve, Bryan Sykes named the originator of this mitochondrial haplogroup Aiyana.
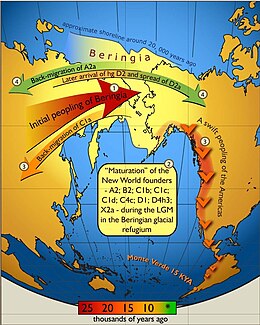
North and South America are continents more recently colonized by modern humans. The migraçãomodelo the hypothesis That by the end of the last Ice Age (about 25.000 years ago) a small number of hunters and gatherers Followed the migration patterns of large animals from northern Asia to North America across the Bering Strait ice bridge. This expansion occurred Rapidly, and pushed to the warmer climates of Central and South America Due to adverse weather conditions and ice That covered most of North America, it is Likely That small scattered groups of Individuals Remained current Alaska, then moved south in the second wave , such as Climatic conditions Gradually improved.
The first four letters of the alphabet are used to designate the four major Native American mtDNAhaplogrupos (A, B, C, and D), and later distinguished from Their Asian counterparts by the addition of a number (A2, B2, C1, D1) . These names refer to four founding lineages That are found in almost all North, Central and South American indigenous populations. A small number of rarer strains have recently Been Identified, but Their Contribution to the overall genetic modern Native American is Likely to Remain minimal. However, the European conquest squeezed native American population through a genetic bottleneck, Reducing Native Americans sets of genes in 1/3 to 1/25 of its size anterior.Esta dramatic change Significantly Reduced genetic variability, and changed forever Genomic groups of survivors. Attempts to reconstruct the genomic structures of most groups of the New World were met with various Difficulties Arising from Lack of this variability.
Genetic analysis of Native American Populations That shows available haplogroup coalescence times range from 16.000 to 22.000 years ago, According to the hypothesis ice bridge.
Native American Haplogroup affiliation and Testing
Although an Increasing number of laboratories offering DNA tests are DNA tests for Native American Y chromosome and mitochondrial DNA, it is important to note That these results are not currently Recognized as sufficient proof to become a registered Native American. Moreover, due to the fact That each of the four major mtDNA haplogroups found in different tribal groups in North America, Central and South America, the Native American DNA test will not Provide a specific tribal affiliation.
Member famous Native American Haplogroups
The Ice Maiden "Juanita" was Discovered on Mount Ampato's Peru, near Arequipa, Peru in 1995 by Johann Reinhard. "Juanita," who lived about 500 years ago, was sacrificed in a religious ceremony around the age of 14 years. Scientists have recovered the DNA of the Ice Maiden and determined That It belonged to the Native American haplogroup A2, and she was Closely related to the Panamanian Ngobe Indian tribe.
References
Torroni A, Schurr TG, et al. Asian Affinities and continental radiation of the four founding Native Americans. J. MtDNAs Human Genetics (1993)
Torroni A, Chen YS, et al. mtDNA and Y chromosome polymorphisms in four indigenous people of southern México.Am. J. Human Genetics (1994).
Bandelt HJ, C. Herrnstadt et al. Identification of Native American Founder mtDNAs Through the analysis of complete mitochondrial DNA sequences:. Some caveats Annals of Human Genetics (2003)
Forster P, Harding R, Torroni A, and Bandelt HJ Origin and evolution of Native American variation mDNA:. The reavaliaçãoAm. J. Human Genetics (1996)
Derenko MV, Grzybowski T, et al. Diversity of mitochondrial DNA lineages in South Siberia. Annals of Human Genetics (2003)
Michael Crawford. The origins of Native Americans: Evidence from Anthropological Genetics, Cambridge University Press (1998)
The first four letters of the alphabet are used to designate the four major Native American mtDNAhaplogrupos (A, B, C, and D), and later distinguished from Their Asian counterparts by the addition of a number (A2, B2, C1, D1) . These names refer to four founding lineages That are found in almost all North, Central and South American indigenous populations. A small number of rarer strains have recently Been Identified, but Their Contribution to the overall genetic modern Native American is Likely to Remain minimal. However, the European conquest squeezed native American population through a genetic bottleneck, Reducing Native Americans sets of genes in 1/3 to 1/25 of its size anterior.Esta dramatic change Significantly Reduced genetic variability, and changed forever Genomic groups of survivors. Attempts to reconstruct the genomic structures of most groups of the New World were met with various Difficulties Arising from Lack of this variability.
Genetic analysis of Native American Populations That shows available haplogroup coalescence times range from 16.000 to 22.000 years ago, According to the hypothesis ice bridge.
Native American Haplogroup affiliation and Testing
Although an Increasing number of laboratories offering DNA tests are DNA tests for Native American Y chromosome and mitochondrial DNA, it is important to note That these results are not currently Recognized as sufficient proof to become a registered Native American. Moreover, due to the fact That each of the four major mtDNA haplogroups found in different tribal groups in North America, Central and South America, the Native American DNA test will not Provide a specific tribal affiliation.
Member famous Native American Haplogroups
The Ice Maiden "Juanita" was Discovered on Mount Ampato's Peru, near Arequipa, Peru in 1995 by Johann Reinhard. "Juanita," who lived about 500 years ago, was sacrificed in a religious ceremony around the age of 14 years. Scientists have recovered the DNA of the Ice Maiden and determined That It belonged to the Native American haplogroup A2, and she was Closely related to the Panamanian Ngobe Indian tribe.
References
Torroni A, Schurr TG, et al. Asian Affinities and continental radiation of the four founding Native Americans. J. MtDNAs Human Genetics (1993)
Torroni A, Chen YS, et al. mtDNA and Y chromosome polymorphisms in four indigenous people of southern México.Am. J. Human Genetics (1994).
Bandelt HJ, C. Herrnstadt et al. Identification of Native American Founder mtDNAs Through the analysis of complete mitochondrial DNA sequences:. Some caveats Annals of Human Genetics (2003)
Forster P, Harding R, Torroni A, and Bandelt HJ Origin and evolution of Native American variation mDNA:. The reavaliaçãoAm. J. Human Genetics (1996)
Derenko MV, Grzybowski T, et al. Diversity of mitochondrial DNA lineages in South Siberia. Annals of Human Genetics (2003)
Michael Crawford. The origins of Native Americans: Evidence from Anthropological Genetics, Cambridge University Press (1998)
http://www.genetree.com/a_and_b
Clan Aiyana is one of 36 groups of mtDNA (or clans) Identified so far. From August 26
2003, Oxford Ancestors searchable database lists 39 games for Aiyana.
According to Oxford Ancestors:
Aiyana
... the founder of one of the four major clans who colonized North America and South East Asia about 12.000 years ago. Descendants Aiyana Fought extreme cold and ice As They crossed the Bering land bridge That joined what is now Alaska and Siberia, to reach the Great Plains of North America. From there, They spread to reach all parts of the continent and, Within just a few thousand years, all of Central America and South America as well. However, Given the data currently available, it is not possible to determine precisely where Aiyana lived or even when.
The descendants of four clans - Chochmingwu, Djigonasee, Aiyana and Ina - dominate the Native Americans. In addition, about 1% of Native Americans are found to belong to the clan of Xenia, Which had its origins on the borders of Europe and Asia. The clans Chochminwu, Djigonasee Aiyana and can be found in modern inhabitants of Siberia and Alaska, but this is not the case of Ina. Their descendants are found in Central and South America, but only the far north Vancouver Island in the northwestern Pacific coast. Interestingly, this is Also the same clan that is Closely Associated with the settlement of the Polynesian islands of Southeast Asia.
The natives of the Far East [Asia] are predominantly descendants of Djigonasee, but there is Considerable representation of "Native Americans" and Aiyana Ina clans, clans and six more - Fufei, Yumi, Nene, Malaxshmi, Emiko and Gaia.
According GeneTree: Cultural History of mtDNA haplogroup A: The origin of Their mtDNA haplotype goes back up to 25.000 years before present, and coincides with the initial peopling of the Americas. The peopling of the Americas occurred with the passage of nomadic Siberian hunters Northeast Asia to Alaska across the Bering Strait. Currently, mtDNA and Y chromosome evidence Suggests That Early migrants to the Americas Originated in Lake Baikal / northern Mongolia. These nomads were great hunters and extremely mobile Rapidly spreading colonized the New World from Alaska to the southern tip of Argentina, in just a few thousand years. The population density Increased and Became the rare big game, these hunters Began to Diversify Their diet and fill all available ecological niches. At the team of European contact, Native Americans lived lifestyles extremely varied, with some groups, such as in Mexico, the development of complex agricultural-based societies and other groups such as the Great Basin, remaining hunter-gatherers, living in small bands furniture .
Posted by ANTONIO on 02/12/2012 12:21:00 PM FLORENTIN
Clan Aiyana is one of 36 groups of mtDNA (or clans) Identified so far. From August 26
2003, Oxford Ancestors searchable database lists 39 games for Aiyana.
According to Oxford Ancestors:
Aiyana
... the founder of one of the four major clans who colonized North America and South East Asia about 12.000 years ago. Descendants Aiyana Fought extreme cold and ice As They crossed the Bering land bridge That joined what is now Alaska and Siberia, to reach the Great Plains of North America. From there, They spread to reach all parts of the continent and, Within just a few thousand years, all of Central America and South America as well. However, Given the data currently available, it is not possible to determine precisely where Aiyana lived or even when.
The descendants of four clans - Chochmingwu, Djigonasee, Aiyana and Ina - dominate the Native Americans. In addition, about 1% of Native Americans are found to belong to the clan of Xenia, Which had its origins on the borders of Europe and Asia. The clans Chochminwu, Djigonasee Aiyana and can be found in modern inhabitants of Siberia and Alaska, but this is not the case of Ina. Their descendants are found in Central and South America, but only the far north Vancouver Island in the northwestern Pacific coast. Interestingly, this is Also the same clan that is Closely Associated with the settlement of the Polynesian islands of Southeast Asia.
The natives of the Far East [Asia] are predominantly descendants of Djigonasee, but there is Considerable representation of "Native Americans" and Aiyana Ina clans, clans and six more - Fufei, Yumi, Nene, Malaxshmi, Emiko and Gaia.
According GeneTree: Cultural History of mtDNA haplogroup A: The origin of Their mtDNA haplotype goes back up to 25.000 years before present, and coincides with the initial peopling of the Americas. The peopling of the Americas occurred with the passage of nomadic Siberian hunters Northeast Asia to Alaska across the Bering Strait. Currently, mtDNA and Y chromosome evidence Suggests That Early migrants to the Americas Originated in Lake Baikal / northern Mongolia. These nomads were great hunters and extremely mobile Rapidly spreading colonized the New World from Alaska to the southern tip of Argentina, in just a few thousand years. The population density Increased and Became the rare big game, these hunters Began to Diversify Their diet and fill all available ecological niches. At the team of European contact, Native Americans lived lifestyles extremely varied, with some groups, such as in Mexico, the development of complex agricultural-based societies and other groups such as the Great Basin, remaining hunter-gatherers, living in small bands furniture .
Posted by ANTONIO on 02/12/2012 12:21:00 PM FLORENTIN
Welcome to
Clan Aiyana
Clan Aiyana
In human genetics, Haplogroup A is a human mitochondrial DNA (mtDNA) haplogroup.
A haplogroup is believed to have originated in Asia about 60,000 years before present. Its ancestral haplogroup was Haplogroup N.
A haplogroup is found throughout modern Asia. Its A1 subgroup is found in northern and central Asia, while its subgroup A2 is found in Siberia and is also one of five haplogroups found nospovos indigenous of the Americas, the others being B, C, D and X.
The mummy "Juanita" from Peru, also called the "Ice Maiden", has been shown to belong to mitochondrial haplogroup A.
In his famous book The Seven Daughters of Eve, Bryan Sykes named the originator of this mitochondrial haplogroup Aiyana.
A haplogroup is believed to have originated in Asia about 60,000 years before present. Its ancestral haplogroup was Haplogroup N.
A haplogroup is found throughout modern Asia. Its A1 subgroup is found in northern and central Asia, while its subgroup A2 is found in Siberia and is also one of five haplogroups found nospovos indigenous of the Americas, the others being B, C, D and X.
The mummy "Juanita" from Peru, also called the "Ice Maiden", has been shown to belong to mitochondrial haplogroup A.
In his famous book The Seven Daughters of Eve, Bryan Sykes named the originator of this mitochondrial haplogroup Aiyana.
North and South America are continents more recently colonized by modern humans. A migraçãomodelo the hypothesis that by the end of the last Ice Age (about 25,000 years ago) a small number of hunters and gatherers followed the migration patterns of large animals from northern Asia to North America across the ice bridge Bering Strait. This expansion occurred rapidly, and pushed to the warmer climates of Central and South America Due to adverse weather conditions and ice that covered most of North America, it is likely that small groups of individuals remained scattered current Alaska, then moved south in a second wave, such as climatic conditions gradually improved.
The first four letters of the alphabet are used to designate the four major Native American mtDNAhaplogrupos (A, B, C, and D), and later Asian distinguished from their counterparts by the addition of a number (A2, B2, C1 , D1). These names refer to four founding lineages that are found in almost all North, Central and South American indigenous populations. A small number of rarer strains have recently been identified, but their contribution to the overall genetic modern Native American is likely to remain minimal. However, the European conquest squeezed native American population through a genetic bottleneck, reducing Native Americans sets of genes in 1/3 to 1/25 of its size anterior.Esta dramatic change significantly reduced genetic variability, and changed forever Genomic groups of survivors. Attempts to reconstruct the genomic structures of most groups of the New World were met with various difficulties arising from this lack of variability.
Genetic analysis of Native American populations shows that coalescence available haplogroup times range from 16,000 to 22,000 years ago, according to the hypothesis ice bridge.
Affiliation Native American Haplogroup and Testing
Although an increasing number of laboratories offering DNA tests are DNA tests for Native American Y chromosome and mitochondrial DNA, it is important to note that these results are not currently recognized as sufficient proof to become a registered Native American. Moreover, due to the fact that each of the four major mtDNA haplogroups found in different tribal groups in North America, Central and South America, a Native American DNA test will not provide a specific tribal affiliation.
Member famous Native American Haplogroups
The Ice Maiden "Juanita" was discovered on Peru's Mount Ampato, near Arequipa, Peru in 1995 by Johann Reinhard. "Juanita," who lived about 500 years ago, was sacrificed in a religious ceremony around the age of 14 years. Scientists have recovered the DNA of the Ice Maiden and determined that it belonged to the Native American haplogroup A2, and she was closely related to the Panamanian Ngobe Indian tribe.
References
Torroni A, Schurr TG, et al. Asian affinities and continental radiation of the four founding Native Americans. J. MtDNAs Human Genetics (1993)
Torroni A, Chen YS, et al. mtDNA and Y chromosome polymorphisms in four indigenous people of southern México.Am. J. Human Genetics (1994).
Bandelt HJ, Herrnstadt C., et al. Identification of Native American Founder mtDNAs Through the analysis of complete mitochondrial DNA sequences:. Some caveats Annals of Human Genetics (2003)
Forster P, Harding R, Torroni A, and Bandelt HJ Origin and evolution of Native American variation mDNA:. A reavaliaçãoAm. J. Human Genetics (1996)
Derenko MV, Grzybowski T, et al. Diversity of mitochondrial DNA lineages in South Siberia. Annals of Human Genetics (2003)
Michael Crawford. The origins of Native Americans: Evidence from Anthropological Genetics, Cambridge University Press (1998)
The first four letters of the alphabet are used to designate the four major Native American mtDNAhaplogrupos (A, B, C, and D), and later Asian distinguished from their counterparts by the addition of a number (A2, B2, C1 , D1). These names refer to four founding lineages that are found in almost all North, Central and South American indigenous populations. A small number of rarer strains have recently been identified, but their contribution to the overall genetic modern Native American is likely to remain minimal. However, the European conquest squeezed native American population through a genetic bottleneck, reducing Native Americans sets of genes in 1/3 to 1/25 of its size anterior.Esta dramatic change significantly reduced genetic variability, and changed forever Genomic groups of survivors. Attempts to reconstruct the genomic structures of most groups of the New World were met with various difficulties arising from this lack of variability.
Genetic analysis of Native American populations shows that coalescence available haplogroup times range from 16,000 to 22,000 years ago, according to the hypothesis ice bridge.
Affiliation Native American Haplogroup and Testing
Although an increasing number of laboratories offering DNA tests are DNA tests for Native American Y chromosome and mitochondrial DNA, it is important to note that these results are not currently recognized as sufficient proof to become a registered Native American. Moreover, due to the fact that each of the four major mtDNA haplogroups found in different tribal groups in North America, Central and South America, a Native American DNA test will not provide a specific tribal affiliation.
Member famous Native American Haplogroups
The Ice Maiden "Juanita" was discovered on Peru's Mount Ampato, near Arequipa, Peru in 1995 by Johann Reinhard. "Juanita," who lived about 500 years ago, was sacrificed in a religious ceremony around the age of 14 years. Scientists have recovered the DNA of the Ice Maiden and determined that it belonged to the Native American haplogroup A2, and she was closely related to the Panamanian Ngobe Indian tribe.
References
Torroni A, Schurr TG, et al. Asian affinities and continental radiation of the four founding Native Americans. J. MtDNAs Human Genetics (1993)
Torroni A, Chen YS, et al. mtDNA and Y chromosome polymorphisms in four indigenous people of southern México.Am. J. Human Genetics (1994).
Bandelt HJ, Herrnstadt C., et al. Identification of Native American Founder mtDNAs Through the analysis of complete mitochondrial DNA sequences:. Some caveats Annals of Human Genetics (2003)
Forster P, Harding R, Torroni A, and Bandelt HJ Origin and evolution of Native American variation mDNA:. A reavaliaçãoAm. J. Human Genetics (1996)
Derenko MV, Grzybowski T, et al. Diversity of mitochondrial DNA lineages in South Siberia. Annals of Human Genetics (2003)
Michael Crawford. The origins of Native Americans: Evidence from Anthropological Genetics, Cambridge University Press (1998)
http://www.genetree.com/a_and_b
Clan Aiyana is one of 36 groups of mtDNA (or clans) identified so far. From August 26
2003, Oxford Ancestors searchable database lists 39 games for Aiyana.
According to Oxford Ancestors:
Aiyana
... the founder of one of the four major clans who colonized North America and South East Asia about 12,000 years ago. Descendants Aiyana fought extreme cold and ice as they crossed the Bering land bridge that joined what is now Alaska and Siberia, to reach the Great Plains of North America. From there, they spread to reach all parts of the continent and, within just a few thousand years, all of Central America and South America as well. However, given the data currently available, it is not possible to determine precisely where Aiyana lived or even when.
The descendants of four clans - Chochmingwu, Djigonasee, Aiyana and Ina - dominate the Native Americans. In addition, about 1% of Native Americans are found to belong to the clan of Xenia, which had its origins on the borders of Europe and Asia. The clans Chochminwu, Djigonasee and Aiyana can be found in modern inhabitants of Siberia and Alaska, but this is not the case of Ina. Their descendants are found in Central and South America, but only as far north as Vancouver Island in northwestern Pacific coast. Interestingly, this is also the same clan that is closely associated with the settlement of the Polynesian islands of Southeast Asia.
The natives of the Far East [Asia] are predominantly descendants of Djigonasee, but there is considerable representation of "Native Americans" and Aiyana Ina clans, clans and six more - Fufei, Yumi, Nene, Malaxshmi, Emiko and Gaia.
According GeneTree: Cultural History of mtDNA haplogroup A: The origin of their mtDNA haplotype goes back up to 25,000 years before present, and coincides with the initial peopling of the Americas. The peopling of the Americas occurred with the passage of nomadic Siberian hunters Northeast Asia to Alaska across the Bering Strait. Currently, mtDNA and Y chromosome evidence suggests that early migrants to the Americas originated in Lake Baikal / northern Mongolia. These nomads were great hunters extremely mobile and rapidly colonized the New World spreading from Alaska to the southern tip of Argentina, in just a few thousand years. As population density increased and became the rare big game, these hunters began to diversify their diet and fill all available ecological niches. At the time of European contact, Native Americans lived lifestyles extremely varied, with some groups, such as in Mexico, the development of complex agricultural-based societies and other groups such as the Great Basin, remaining hunter-gatherers, living in small bands
Clan Aiyana is one of 36 groups of mtDNA (or clans) identified so far. From August 26
2003, Oxford Ancestors searchable database lists 39 games for Aiyana.
According to Oxford Ancestors:
Aiyana
... the founder of one of the four major clans who colonized North America and South East Asia about 12,000 years ago. Descendants Aiyana fought extreme cold and ice as they crossed the Bering land bridge that joined what is now Alaska and Siberia, to reach the Great Plains of North America. From there, they spread to reach all parts of the continent and, within just a few thousand years, all of Central America and South America as well. However, given the data currently available, it is not possible to determine precisely where Aiyana lived or even when.
The descendants of four clans - Chochmingwu, Djigonasee, Aiyana and Ina - dominate the Native Americans. In addition, about 1% of Native Americans are found to belong to the clan of Xenia, which had its origins on the borders of Europe and Asia. The clans Chochminwu, Djigonasee and Aiyana can be found in modern inhabitants of Siberia and Alaska, but this is not the case of Ina. Their descendants are found in Central and South America, but only as far north as Vancouver Island in northwestern Pacific coast. Interestingly, this is also the same clan that is closely associated with the settlement of the Polynesian islands of Southeast Asia.
The natives of the Far East [Asia] are predominantly descendants of Djigonasee, but there is considerable representation of "Native Americans" and Aiyana Ina clans, clans and six more - Fufei, Yumi, Nene, Malaxshmi, Emiko and Gaia.
According GeneTree: Cultural History of mtDNA haplogroup A: The origin of their mtDNA haplotype goes back up to 25,000 years before present, and coincides with the initial peopling of the Americas. The peopling of the Americas occurred with the passage of nomadic Siberian hunters Northeast Asia to Alaska across the Bering Strait. Currently, mtDNA and Y chromosome evidence suggests that early migrants to the Americas originated in Lake Baikal / northern Mongolia. These nomads were great hunters extremely mobile and rapidly colonized the New World spreading from Alaska to the southern tip of Argentina, in just a few thousand years. As population density increased and became the rare big game, these hunters began to diversify their diet and fill all available ecological niches. At the time of European contact, Native Americans lived lifestyles extremely varied, with some groups, such as in Mexico, the development of complex agricultural-based societies and other groups such as the Great Basin, remaining hunter-gatherers, living in small bands
Genetic history of indigenous peoples of the Americas
From Wikipedia, the free encyclopedia
For a non-technical introduction to genetics in general, see Introduction to genetics.

An autosomal genetic tree showing some neighbour-joining relationships within Amerindian peoples
Genetic history of indigenous peoples of the Americas primarily focus on Human Y-chromosome DNA haplogroups andHuman mitochondrial DNA haplogroups.[1] Autosomal "atDNA" markers are also used, but differ from mtDNA or Y-DNA in that they overlap significantly.[2] The genetic pattern indicates Indigenous Amerindians experienced two very distinctive genetic episodes; first with the initial peopling of the Americas, and secondly with European colonization of the Americas.[3][4] The former is the determinant factor for the number of gene lineages, zygosity mutations and founding haplotypes present in today's Indigenous Amerindian populations.[3]
Human settlement of the New World occurred in stages from the Bering sea coast line, with an initial layover on Beringia for the small founding population.[5][6][7] The micro-satellite diversity and distributions of the Y lineage specific to South Americaindicates that certain Amerindian populations have been isolated since the initial colonization of the region.[8] The Na-Dené,Inuit and Indigenous Alaskan populations exhibit haplogroup Q (Y-DNA); however, they are distinct from other indigenous Amerindians with various mtDNA and atDNA mutations.[9][10][11] This suggests that the peoples who first settled the northern extremes of North America and Greenland derived from later migrant populations than those who penetrated further south in the Americas.[12][13] Linguists and biologists have reached a similar conclusion based on analysis of Amerindian language groups and ABO blood group system distributions.[14][15][16]
Contents
[hide]
Background[edit]
Further information: Timeline of human evolution and Recent African origin of modern humans

A single-nucleotide polymorphism (SNP) is a change to a single nucleotide in a DNA sequence.
The Y chromosome is passed down directly from father to son; all male humans (Y chromosomes) today trace back to a single prehistoric father termed "Y chromosomal Adam" originating in Africa.[17] The Y chromosome spans about 60 million base pairs (the building blocks ofDNA) and represents about 2 percent of the total DNA in all human cells.[18] The original "Y chromosomal Adam"-DNA sequencing has mutated rarely over the 20,000 generations, but each time a new mutation occurs, there is a new branch in a haplogroup, resulting in a newsubclade (single-nucleotide polymorphism (SNP)).[19]
MtDNA mutations are also passed down relatively unchanged from generation to generation, so all humans share the same mtDNA-types. The logical extension of this is that all humans ultimately trace back to one woman, who is commonly referred to as Mitochondrial Eve.[20][21]Both females and males inherit their Mitochondrial DNA (mtDNA) only from their mother.[22] This line of biological inheritance, therefore, stops with each male.[23] Consequently, Y-DNA is more commonly used by the general public for tracing genetic heritage of a direct male line.[23][24][25]
An autosome (atDNA) is a chromosome that is not a sex chromosome – that is to say, there are an equal number of copies of the chromosome in males and females.[2] Autosomal DNA testing is generally used to determine the "genetic percentages" of a person's ancestry from particular continents/regions or to identify the countries and "tribes" of origin on an overall basis. Genetic admixture tests arrive at these percentages by examining (SNP), which are locations on the DNA where one nucleotide has "mutated" or "switched" to a differentnucleotide.[2] One way to examine the support for particular colonization routes within the American landmass is to determine if a closer relationship between zygosity and geography is observed when “effective” geographic distances are computed along these routes, rather than along shortest-distance paths.[26]
Y-DNA[edit]
- For more details on individual Amerindian groups by Y-DNA, see Y-DNA haplogroups in Indigenous peoples of the Americas.
The Y chromosome consortium has established a system of defining Y-DNA haplogroups by letters A through to T, with further subdivisions using numbers and lower case letters.[27]
Haplogroup Q[edit]
For more details on this topic, see Haplogroup Q-M242 (Y-DNA).
Q-M242 (mutational name) is the defining (SNP) of Haplogroup Q (Y-DNA) (phylogenetic name). Within the Q clade, there are 14 haplogroups marked by 17 SNPs.2009[28][29] In Eurasia haplogroup Q is found among indigenousSiberian populations, such as the modern Chukchi and Koryak peoples. In particular, two groups exhibit large concentrations of the Q-M242 mutation, the Ket (93.8%) and the Selkup (66.4%) peoples.[30] The Ket are thought to be the only survivors of ancient wanderers living in Siberia.[5] Their population size is very small; there are fewer than 1,500 Ket in Russia.2002[5] The Selkup have a slightly larger population size than the Ket, with approximately 4,250 individuals.[31]
Starting the Paleo-Indians period, a migration to the Americas across the Bering Strait (Beringia) by a small population carrying the Q-M242 mutation took place.[32] A member of this initial population underwent a mutation, which defines its descendant population, known by the Q-M3 (SNP) mutation.[33] These descendants migrated all over the Americas.[28]
Haplogroup Q-M3 (Y-DNA) is defined by the presence of the rs3894 (M3) (SNP).[3][5][34] The Q-M3 mutation is roughly 15,000 years old as that is when the initial migration of Paleo-Indians into the Americas occurred.[7][35] Q-M3 is the predominant haplotype in the Americas, at a rate of 83% in South American populations,[36] 50% in the Na-Dené populations, and in North American Eskimo-Aleut populations at about 46%.[30] With minimal back-migration of Q-M3 in Eurasia, the mutation likely evolved in east-Beringia, or more specifically the Seward Peninsula or western Alaskan interior. The Beringia land mass began submerging, cutting off land routes.[6][30][37]
Since the discovery of Q-M3, several subclades of M3-bearing populations have been discovered. An example is in South America, where some populations have a high prevalence of (SNP) M19 which defines subclade Q-M19.[36] M19 has been detected in (59%) of Amazonian Ticuna men and in (10%) of Wayuu men.[36] Subclade M19 appears to be unique to South American Indigenous peoples, arising 5,000 to 10,000 years ago.[36] This suggests that population isolation and perhaps even the establishment of tribal groups began soon after migration into the South American areas.[5][38] Other American subclades include Q-L54, Q-Z780, Q-MEH2, Q-SA01, and Q-M346 lineages. In Canada, two other lineages have been found. These are Q-P89.1 and Q-NWT01.
Haplogroup R1[edit]
For more details on this topic, see Haplogroup R1 (Y-DNA).
Haplogroup R1 (Y-DNA) (specially R1b) is the second most predominant Y haplotype found among indigenous Amerindians after Q (Y-DNA).[39] The distribution of R1 is believed to be associated with the re-settlement of Eurasia following the last glacial maximum. One theory put forth is that it entered the Americas with the initial founding population.[40] A second theory is that it was introduced during European colonization.[39] R1 is very common throughout all of Eurasia except East Asia and Southeast Asia. R1 (M173) is found predominantly in North American groups like the Ojibwe (79%), Chipewyan (62%), Seminole (50%), Cherokee (47%), Dogrib (40%) andPapago (38%).[39]
Haplogroup C-P39[edit]
For more details on this topic, see Haplogroup C-M217 (Y-DNA).
Haplogroup C-M217 is mainly found in indigenous Siberians, Mongolians and Oceanic populations. Haplogroup C-M217 is the most widespread and frequently occurring branch of the greater (Y-DNA) haplogroup C-M130. Haplogroup C-M217 descendant C-P39 is commonly found in today's Na-Dené speakers, with the highest frequency found among the Athabaskans at 42%.[32] This distinct and isolated branch C-P39 includes almost all the Haplogroup C-M217 Y-chromosomes found among all indigenous peoples of the Americas.[41] The Na-Dené groups are also unusual among indigenous peoples of the Americas in having a relatively high frequency of Q-M242 (25%).[30]
Some researcher feel that this may indicate that the Na-Dené migration occurred from the Russian Far East after the initial Paleo-Indian colonization, but prior to modern Inuit, Inupiat and Yupik expansions.[9][10][42]
mtDNA[edit]
For more details on mtDNA in general, see Mitochondrial DNA.
Mitochondrial Eve is defined as the woman who was the matrilineal most recent common ancestor for all living humans. Mitochondrial Eve is estimated to have lived between 140,000 and 200,000 years ago,[43][44] Mitochondrial Eve is the most recent common matrilineal ancestor, not the most recent common ancestor.[45][46]
When studying human mitochondrial DNA (mtDNA) haplogroups, the results indicated until recently that Indigenous Amerindian haplogroups, including haplogroup X, are part of a single founding east Asian population. It also indicates that the distribution of mtDNA haplogroups and the levels of sequence divergence among linguistically similar groups were the result of multiple preceding migrations from Bering Straits populations.[22][47] All Indigenous Amerindian mtDNA can be traced back to five haplogroups, A, B, C,D and X.[48] More specifically, indigenous Amerindian mtDNA belongs to sub-haplogroups that are unique to the Americas and not found in Asia or Europe: A2, B2, C1, D1, and X2a (with minor groups C4c, D2, D3, and D4h3).[47] This suggests that 95% of Indigenous Amerindian mtDNA is descended from a minimal genetic founding female population, comprising sub-haplogroups A2, B2, C1b, C1c, C1d, and D1.[48] The remaining 5% is composed of the X2a, D2, D3, C4, and D4h3 sub-haplogroups.[47][48]
X is one of the five mtDNA haplogroups found in Indigenous Amerindian peoples. Unlike the four main American mtDNA haplogroups (A, B, C and D), X is not at all strongly associated with east Asia.[5] Haplogroup X genetic sequences diverged about 20,000 to 30,000 years ago to give two sub-groups, X1 and X2. X2's subclade X2a occurs only at a frequency of about 3% for the total current indigenous population of the Americas.[5] However, X2a is a major mtDNA subclade in North America; among theAlgonquian peoples, it comprises up to 25% of mtDNA types.[49][50] It is also present in lower percentages to the west and south of this area — among the Sioux (15%), the Nuu-chah-nulth (11%–13%), the Navajo (7%), and the Yakama (5%).[51] Haplogroup X is more strongly present in the Near East, the Caucasus, and Mediterranean Europe.[51] The predominant theory for sub-haplogroup X2a's appearance in North America is migration along with A, B, C, and D mtDNA groups, from a source in the Altai Mountains of central Asia.[52][53][54][55]
Sequencing of the mitochondrial genome from Paleo-Eskimo remains (3,500 years old) are distinct from modern Amerindians, falling within sub-haplogroup D2a1, a group observed among today's Aleutian Islanders, the Aleut and Siberian Yupikpopulations.[56] This suggests that the colonizers of the far north, and subsequently Greenland, originated from later coastal populations.[56] Then a genetic exchange in the northern extremes introduced by the Thule people (proto-Inuit) approximately 800–1,000 years ago began.[11][57] These final Pre-Columbian migrants introduced haplogroups A2a and A2b to the existing Paleo-Eskimo populations of Canada and Greenland, culminating in the modern Inuit.[11][57]
A 2013 study in Nature reported that DNA found in the 24,000-year-old remains of a young boy from the archaeological Mal'ta-Buret' culture suggest that up to one-third of the indigenous Americans may have ancestry that can be traced back to western Eurasians, who may have "had a more north-easterly distribution 24,000 years ago than commonly thought"[58] "We estimate that 14 to 38 percent of Native American ancestry may originate through gene flow from this ancient population," the authors wrote. Professor Kelly Graf said,
"Our findings are significant at two levels. First, it shows that Upper Paleolithic Siberians came from a cosmopolitan population of early modern humans that spread out of Africa to Europe and Central and South Asia. Second, Paleoindian skeletons like Buhl Woman with phenotypic traits atypical of modern-day indigenous Americans can be explained as having a direct historical connection to Upper Paleolithic Siberia."
A route through Beringia is seen as more likely than the Solutrean hypothesis.[59] An abstract in a 2012 issue of the "American Journal of Physical Anthropology" states that "The similarities in ages and geographical distributions for C4c and the previously analyzed X2a lineage provide support to the scenario of a dual origin for Paleo-Indians. Taking into account that C4c is deeply rooted in the Asian portion of the mtDNA phylogeny and is indubitably of Asian origin, the finding that C4c and X2a are characterized by parallel genetic histories definitively dismisses the controversial hypothesis of an Atlantic glacial entry route into North America."[60]
AtDNA[edit]
For more details on autosomal DNA in general, see Human genetic variation.

genetic groups according to DNA Tribes
1. Arctic
2. Salishan
3. Athabaskan
4. North Amerindian
5. Ojibwa
6. Mexica
|
7. Maya
9. Andean
10. Amazonian
11. Gran Chaco
12. Patagonian
|
Genetic diversity and population structure in the American landmass is also done using autosomal (atDNA) micro-satellite markersgenotyped; sampled from North, Central, and South America and analyzed against similar data available from other indigenous populations worldwide.[26][61] The Amerindian populations show a lower genetic diversity than populations from other continental regions.[61] Observed is a decreasing genetic diversity as geographic distance from the Bering Strait occurs, as well as a decreasing genetic similarity to Siberian populations from Alaska (the genetic entry point).[26][61]
Also observed is evidence of a higher level of diversity and lower level of population structure in western South America compared to eastern South America.[26][61] There is a relative lack of differentiation between Mesoamerican and Andean populations, a scenario that implies that coastal routes were easier for migrating peoples (more genetic contributors) to traverse in comparison with inland routes.[26]
The over-all pattern that is emerging suggests that the Americas were colonized by a small number of individuals (effective size of about 70), which grew by a factor of 10 over 800 – 1000 years.[5][62] The data also shows that there have been genetic exchanges between Asia, the Arctic, and Greenland since the initial peopling of the Americas.[62][63]
In 2014, the autosomal DNA of a 12,500+-year-old infant from Montana was sequenced.[64] The DNA was taken from a skeleton referred to as Anzick-1, found in close association with several Clovis artifacts. Comparisons showed strong affinities with DNA from Siberian sites, and virtually ruled out any close affinity with European sources (the so-called "Solutrean hypothesis"). The DNA also showed strong affinities with all existing Native American populations, which indicated that all of them derive from an ancient population that lived in or near Siberia, the Upper Palaeolithic Mal'ta population.[65]
Overlaps between DNA types[edit]
Further information: Population genetics and Y-DNA haplogroups in Indigenous peoples of the Americas
Populations that have a specific combination of autosome, Y and MT-haplogroup mutations can generally be found with regional variations. Autosomes, Y mutations and mt mutations do not necessarily occur at a similar time and there are differential rates of sexual selection between the two sex chromosomes.[66] This combined with population bottlenecks, the founder effect, mitochondrial mutations and genetic drift will alter the genetic composition of isolated populations, resulting in very distinguishable mutation patterns.[67] (i.e. Taínos,[68] Fuegians,[69] Inuit,[70] Yupik[71] and Algonquian[5])
The rough overlaps between Y-DNA and mtDNA between the Americas, Circumpolar north, and Siberian indigenous populations are:
| Y-DNA haplogroup(s) - | mtDNA haplogroup(s) - | Geographical area(s) |
|---|---|---|
| Q-M242, R1, C-M217 | A, X, Y, C, D (M types), (N types) | Russian far east, Americas, Arctic |
Old world genetic admixture[edit]
For more details on genetic admixture in the Americas, see Miscegenation § Genetic studies of racial admixture.
See also: Multiracial American

"Las castas" - Painting containing complete set of 16casta combinations. An 18th century socio-racial classification system used in the Spanish American colonies.
Interracial marriage and interracial sex and, more generally, the process of racial admixture, has its origins in prehistory. Racial mixing became widespread during European colonialism in the Age of Discovery.[72] Genetic exchange between two populations reduces the genetic distancebetween the populations and is measurable in DNA patterns.[66] During the Age of Discovery, beginning in the late 1400s, European explorers sailed the oceans, eventually reaching all the major continents.[73] During this time Europeans contacted many populations, some of which had been relatively isolated for millennia.[74] The genetic demographic composition of the Eastern Hemisphere has not changed significantly since the age of discovery. However, genetic demographics in the Western Hemisphere were radically altered by events following the voyages ofChristopher Columbus.[74] The European colonization of the Americas brought contact between peoples of Europe, Africa and Asia and the Amerindian populations. As a result, the Americas today have significant and complex multiracial populations.[74] Many individuals who self-identify as one race exhibit genetic evidence of a multiracial ancestry.[75]
The European conquest of Latin America beginning in the late 15th century, was initially executed by male soldiers and sailors from the Iberian Peninsula (Spain and Portugal).[76] The new soldier-settlers fathered children with Amerindian women and later with African slaves.[77] Thesemixed-race children were generally identified by the Spanish colonist and Portuguese colonist as "Castas".[78] The subsequent North American fur trade during the 16th century brought many more European men, from France, Ireland, and Great Britain, who took North Amerindian women as wives.[79] Their children became known as "Métis" or "Bois-Brûlés" by the French colonists and "mixed-bloods", "half-breeds" or "country-born" by the English colonists and Scottish colonists.[80] From the second half of the 19th century to the beginning of the 20th century, new waves of immigrants from northern, eastern and southern Europe went to the Americas and consequently altered the demographics.[81] Following World War II and subsequent worldwide migrations, the current American populations' genetic admixture can be traced to all corners of the world.[5][82]
Blood groups[edit]
Further information: ABO blood group system
Prior to the 1952 confirmation of DNA as the hereditary material by Alfred Hershey and Martha Chase, scientists usedblood proteins to study human genetic variation.[83][84] The ABO blood group system is widely credited to have been discovered by the Austrian Karl Landsteiner, who found three different blood types in 1900.[85] Blood groups are inherited from both parents. The ABO blood type is controlled by a single gene (the ABO gene) with three alleles: i,IA, and IB.[86]
Research by Ludwik and Hanka Herschfeld during World War I found that the frequencies of blood groups A,B and O differed greatly from region to region.[84] The "O" blood type (usually resulting from the absence of both A and B alleles) is very common around the world, with a rate of 63% in all human populations.[87] Type "O" is the primary blood type among the indigenous populations of the Americas, in-particular within Central and South America populations, with a frequency of nearly 100%.[87] In indigenous North American populations the frequency of type "A" ranges from 16% to 82%.[87] This suggests again that the initial Amerindians evolved from an isolated population with a minimal number of individuals.[88][89]
| PEOPLE GROUP | O (%) | A (%) | B (%) | AB (%) |
|---|---|---|---|---|
| Blackfoot (N. Am. Indian) | 17 | 82 | 0 | 1 |
| Bororo (Brazil) | 100 | 0 | 0 | 0 |
| Eskimos (Alaska) | 38 | 44 | 13 | 5 |
| Inuit (Eastern Canada & Greenland) | 54 | 36 | 23 | 8 |
| Hawaiians | 37 | 61 | 2 | 1 |
| Indigenous North Americans (as a whole Native Nations/First Nations) | 79 | 16 | 4 | 1 |
| Maya (modern) | 98 | 1 | 1 | 1 |
| Navajo | 73 | 27 | 0 | 0 |
| Peru | 100 | 0 | 0 | 0 |
Genealogical testing[edit]
Main article: Genealogical DNA test
A genealogical DNA test examines the nucleotides at specific locations on a person's DNA for genetic genealogy purposes.[91] The test results are not meant to have any medical value; they are intended only to give genealogical information. Genealogical DNA tests generally involve comparing the results of living individuals to historic populations.[91] The general procedure for taking a genealogical DNA test involves taking a painless cheek-scraping (also known as a buccal swab) at home and mailing the sample to a genetic genealogy laboratory for testing. Genetic testing results showing specific sub-Haplogroups of Q, R1 and C3b implies that the individuals ancestry is, in whole or in-part, indigenous to the Americas.[92] If one's mtDNA belonged to specific sub-Haplogroups of, A, B, C, D or X2a, the implication would be that the individual's ancestry is, in whole or part, indigenous to the Americas.[93]
See also[edit]
.
Postado por ANTONIO FLORENTINO às 12/02/2012 12:21:00 PM











Nenhum comentário:
Postar um comentário